In this study, we wanted to get new insights into breast cancer heterogeneity by establishing metabolic clusters of breast cancer and further explore these with available gene and protein data. Sample material from 228 primary operable patients were analyzed with high-resolution magic angle spinning MR spectroscopy (HR MAS MRS) to extract metabolic information. Additionally, 201 of the samples had been analyzed by microarray to get gene expression data and 217 of them with reverse phase protein array (RPPA) to extract protein data.
We discovered three clusters (Figure 1) with significant differences in glucose and choline metabolism, both of which are pathways known to be aberrant in cancer. Therapeutic agents that target metabolic dependencies are considered to be a promising anti-cancer strategy, and the metabolic differences of the clusters could have future relevance for treatment stratification. On the protein level (RPPA subtypes), the clusters expressed differences in breast cancer related proteins. However, only one of the clusters was significantly different when considering gene expressions. Importantly, this cluster showed differences in the expression of genes and proteins related to the extracellular matrix. The gene expression subtypes (PAM50) were distributed evenly among the clusters, indicating that the metabolic clusters might add information for understanding breast cancer heterogeneity.
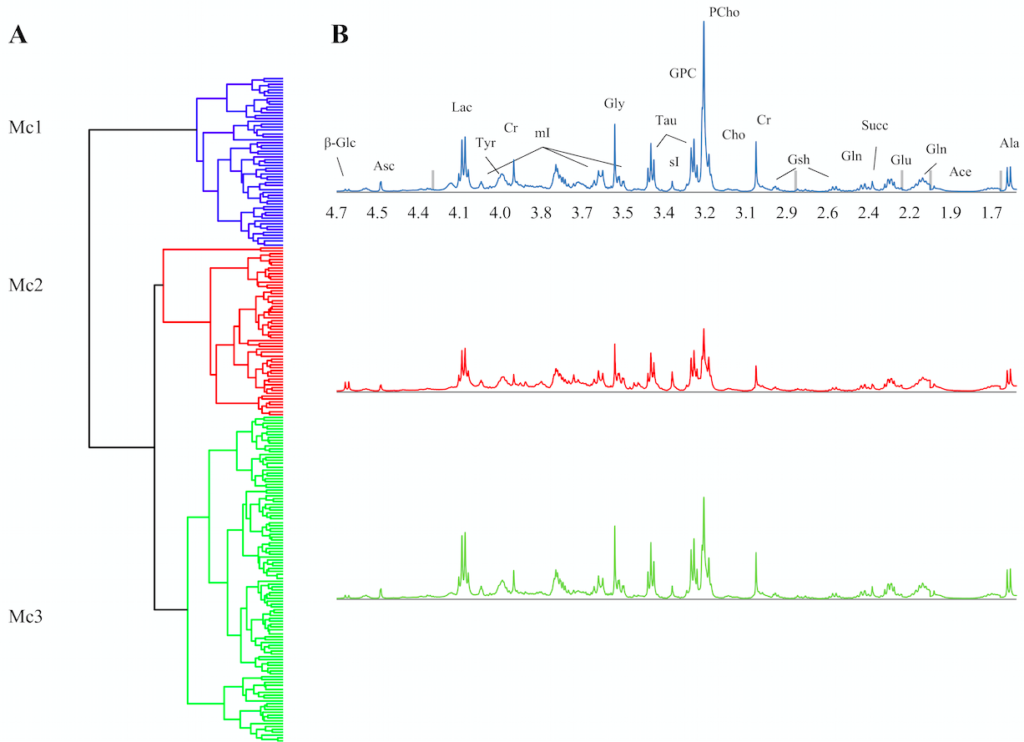
Figure 1: The samples were divided into three clusters based on differences in their metabolic profiles from MR spectroscopy.
The paper was published in Cancer and Metabolism.