Authors: Leslie Euceda, Scientist, and Debbie Hill, Senior engineer.
Breast cancer is one of the major topics we investigate at the MR Cancer group. There are different substances inside breast cancer cells that can tell us different things. For example, if receptors for the hormones estrogen and progesterone are present in a breast tumor, the patient will probably respond to hormonal therapy. Similarly, some breast cancers have large amounts of a protein called HER2, which pushes cancer cells to multiply uncontrollably; these patients have a high chance of responding to anti-HER2 therapy. But then there are the breast cancers that don’t display any of these features. These are classified as triple negative breast cancers; they do not respond to hormonal therapy or anti-HER2 therapy. Tumors from these patients will usually be surgically removed, but the follow-up treatment is limited to classic chemotherapy, which is more aggressive and since it affects the entire body, gives more side effects. Even with this treatment, triple negative breast cancer patients have a high risk of the cancer coming back.
Many scientists and doctors around the world are working to find better treatments for triple negative breast cancer. A potential drug is everolimus. Everolimus blocks a particular cellular process that is known to be especially active in triple negative breast cancer. Initial tests of everolimus in mouse models of triple negative breast cancer have been somewhat successful, but not all cases respond. Our aim was to see if we could predict triple negative breast cancer patient response to everolimus treatment.
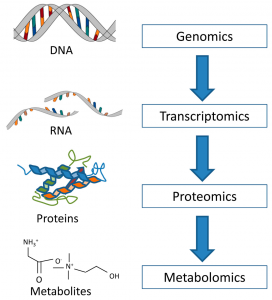
In order to predict patient response, we first need to understand how the cells are organized. Cells contain our biological information structured in four levels. The genes, which contain the instructions for any given situation in the cell, are on the first level. Triggered in response to a particular situation, certain genes are expressed; they stand out from the rest, sending a message about the course of action. The expressed genes are RNA, which make up the second level. The employees of the cell, or proteins, are on the third level. They are activated or synthesized on instruction by the RNA. The hormone receptors and the HER2 protein are examples of proteins. Finally, while the proteins do their work, they will produce different materials. Some of these can be useful while others are just waste. These materials are what we call metabolites; they are on the fourth and last level.
Being the last level of biological information, metabolites are closer to the actual observable characteristics of the cell. These are influenced by external factors like environment, exercise, and diet. So, metabolites can give different information than the other levels. For example, there may be nothing wrong with the instructions (genes) but the employees (proteins) may be sick. Or the employees may be fine but the materials may be expired. So, we wondered whether measuring metabolites might add information that can help predict response to everolimus.
We measured metabolites in over 100 triple negative breast tumors using nuclear magnetic resonance (NMR) spectroscopy [1]. Tumor samples were taken from triple negative breast cancer patients, and then grown in mice to preserve the human tumor characteristics. Then, we treated some tumors with everolimus and left others untreated, which allowed us to investigate the effect of everolimus under controlled conditions, which would not be possible with human patients. We found clear metabolite differences between the tumors that were treated with everolimus and those that were not, which means that everolimus had an effect on tumor metabolism. Unfortunately, we could not predict which mice responded well to everolimus based only on the metabolites we measured. Still, we found that the way the metabolites change with everolimus treatment is different between the responding and non-responding mice. The differences we found suggest that the metabolic response to everolimus treatment is bigger in the responders.
Previous studies at the MR Cancer group have shown that triple negative breast tumors in mice have different metabolite characteristics compared to other breast cancer subtypes with a better prognosis [2] and that the way these subtypes change with treatment is different [3]. We are now investigating if we can see the same thing in triple negative breast tumors from human patients.
Ultimately, the goal of our work is to improve cancer treatment and patient care, and we could not do it without funding from organizations like the Research Council of Norway (Forskningsrådet), the Norwegian Cancer Society (Kreftforeningen) and the the Liaison Committee between Central Norway RHA and NTNU.
If you’re feeling just a little inclined to help the cause, don’t hesitate to get involved! Whether it’s just by wearing pink or participating in events near you that support the fight against breast cancer, let yourself get caught by the October breast cancer awareness spirit!
This text was first published at NTNU Medisin og Helse Fagblogg.
References
- Euceda LR, Hill DK, Stokke E, Hatem R, El Botty R, Bieche I et al. Metabolic Response to Everolimus in Patient-Derived Triple-Negative Breast Cancer Xenografts. J Proteome Res. 2017;16(5):1868-79. doi:10.1021/acs.jproteome.6b00918.
- Grinde M, Skrbo N, Moestue S, Rødland E, Borgan E, Kristian A, et al. Interplay of choline metabolites and genes in patient-derived breast cancer xenografts. Breast Cancer Res. 2014;16:1-16.
- Moestue SA, Dam CG, Gorad SS, Kristian A, Bofin A, Mælandsmo GM et al. Metabolic biomarkers for response to PI3K inhibition in basal-like breast cancer. Breast Cancer Res. 2013;15:R16.